Home »Unlabelled » Computers designed for Proteomics processing!
Friday, August 8, 2014
Computers designed for Proteomics processing!
It was just a matter of time before someone did this, right? I get questions about this through the blog and through my day job all the time. The truth is it isn't easy to figure out what is going to make a good processing computer. You'd think it would be simpler, right? Old computers will be slow, new computers will be fast, and expensive new computers will be the fastest. Yet I've been in labs this summer that have dumped $3,000 + on Xeon desktops that are much slower than my quad core laptop I'm always bragging about. To this day, I definitely can not guess why one PC is going to be slower than another.
Fortunately, I don't have to worry about it anymore. These guys at OmicsComputing have put together 2 processing PCs, a basic "Omics Workstation" for fast processing and a super processing computer called, get this, the "Proteome Destroyer".
If you've read much on my blog, you know what a dork I am for stuff like this. So my first thought was to see if I could borrow some time on one of these and run some stuff. And I got to. And they aren't messing around.
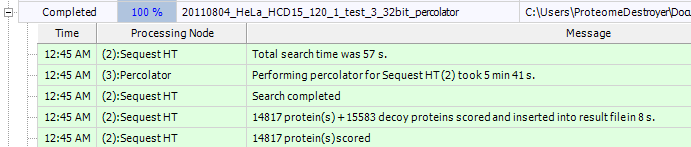
Sorry the text is small here. What is it? My favorite HeLa high high file ran on the Proteome Discoverer 1.4 demo. I used the full human Uniprot. Static modification of alkylated cysteines and dynamic mod of oxidized methionine. Sequest took 57 seconds and Percolator (32-bit, not the awesome beta that I have accidentally shown you guys before) took under 6 minutes. So...a whole HeLa high-high run in 7 minutes or so. Pretty good. I've done better on my overclocked quad core with the Percolator beta we're testing, but this beats the heck out of virtually every other run I've seen.
Okay, who cares, high high files are easy. What about those big Fusion files that even my quad core suffers in processing (we're talking High/low and hugely dense data files) 20 minutes, with Percolator (misplaced the screenshot for proof, but I'll load it later. I know its in one of these inboxes somewhere...). I have never ever processed a Fusion HeLa file in under 30 minutes before....
Get this, though, apparently this isn't nearly as fast as this PC can go. When I reported back what I saw (probably faster than I've seen, but not mind blowing) they took a closer look at the runs and the processor wasn't running anywhere near maximum. I guess it uses a very aggressive processing boost function when it is under high load. Proteome Discoverer running a Fusion file wasn't enough to trigger high load functioning. The PC was like, "oh well, I'll run this but I don't need to activate all of my cores or memory or anything."
So they tweaked the software or hardware or something so that it recognizes PD as a software that should be ran at full capacity and invited me to try re-running my files. As you might guess, I'm psyched to test it out! As you also might guess, it may be a while till I can get to it cause I've got lots of other things to do.
You can check out their simple little webstore here.
TL/DR: This company designs computers just for genomics and proteomics processing and I'm pretty sure they are a whole lot faster than what you are processing your Proteome Discover data with. And apparently, I didn't see the full capacity of what these computers can do! BTW, they aren't nearly as expensive as you'd think, as they run from $1800 - $4000, crazy, right!?!?!
Subscribe to:
Post Comments (Atom)

0 comments:
Post a Comment